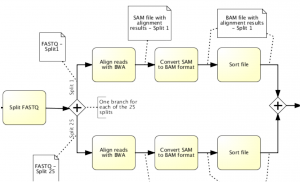
 Raw genome data acquired from next-generation sequencing devices need to be preprocessed prior its analysis. This involves steps for reconstruction or alignment of the whole genome and the identification of variants compared to a reference or variant calling. However, there are various tuning points, such as quality control, filtering, or realignment, in the process, which makes it hard to have a single genome data processing pipeline for all purposes.
Raw genome data acquired from next-generation sequencing devices need to be preprocessed prior its analysis. This involves steps for reconstruction or alignment of the whole genome and the identification of variants compared to a reference or variant calling. However, there are various tuning points, such as quality control, filtering, or realignment, in the process, which makes it hard to have a single genome data processing pipeline for all purposes.
Design Time
We integrated the Business Process Modeling and Notation (BPMN) to enable graphical model, adaption, and configuration of pipelines. During design time, you can use the modeling environment to create your own pipeline models. If you need assistance, we can provide modeling as a service for you. Pipeline models can be parametrized, i.e. you can specify tuning points during design time, which need to be set during configuration. Parameters are an easy way to have a variety of similar pipelines without the need to adapt the model itself.
Configuration Time
After selecting a specific pipeline model, the configuration of a pipeline model instance starts. During configuration all parameters of the model need to be configured with concrete values. For example, if you specified a pipeline model with a process step placement for alignment, the concrete alignment algorithm is set during configuration. Furthermore, you can also set tool specific configuration parameters, such as minimum quality for variant calling during configuration of a pipeline model instance.
Execution Time
The execution of the created pipeline model instance is the final step. In the task control center you select the appropriate pipeline model instance and its input files, such as raw genome data in FASTQ files. Since all process specific parameters were set during configuration time, the execution time requires only a minimum of parameters before you can start the execution of the pipeline model instance. This is designed to save time during the processing of your acquired data. For example, consider you want to process hundreds of files of an experiment, which requires the same genome data processing pipeline instance. In this case, you only need to select all input files and the appropriate pipeline model instance and start its execution. The coordination of individual tasks and the processing using your modeled pipeline is supervised by the management system.
Available Pipelines
We have pre-assembled a set of ready-to-use pipelines for you allowing analysis of both single- and paired-read sequencing data. All offer you a particular range of flexibility in choosing tools for the distinct processing steps.
Standard (Single-Read)
This pipeline follows the standard analysis procedure involving tool-based splitting, sorting, indexing, and merging that are necessary for general and distributed processing. For those intermediate steps we apply SAMtools. For alignment, users can choose from a range of tools, e.g. BWA, HANA Alignment, or Bowtie. Variant calling is conducted with bcftools.
Optimized (Single-Read)
This type of pipeline makes use of in-memory database technology to accelerate data processing. It can apply the same analysis tools for alignment and variant calling as the Standard pipeline. However, formerly mandatory intermediate processing steps like splitting, sorting, and merging are not executed by dedicated tools anymore but handled by our computing platform. Pipelines of this type usually drastically speed up processing time compared to Standard pipelines.
Standard (Paired-Read)
This type of pipeline conducts alignment with BWAMem and variant calling with GATK’s UnifiedGenotyper. In addition, it applies all intermediate steps to improve data quality as recommended by GATK’s best practices. Those steps include read duplicate detection, local realignment, and Base Quality Score Recalibration (BQSR).
Simple (Paired-Read)
This type of pipeline conducts alignment with BWAMem and variant calling with GATK’s UnifiedGenotyper without any intermediate steps for improving data quality, e.g. BQSR.
Amplicon (Paired-Read)
This type of pipeline is supposed to be used for targeted sequencing data. It applies the same steps as the Standard pipeline for paired reads, but uses only a particular set of genes as reference.
Available Tools
Burrows-Wheeler-Aligner (BWA)
Burrows-Wheeler-Aligner (BWA) is a software package for mapping next-generation sequencing data to a reference genome. To reduce the memory footprint of the reference index, BWA algorithms apply Burrows-Wheeler transform. From this software package, we apply BWA-backtrack and BWA-MEM. The former is designed for reads of up to 100bp length and available for our single-read pipelines, whilst the latter is available for paired-read pipelines, suitable for read sequences ranging from 70bp to 1Mbp, and generally faster and more accurate.
Torrent Mapping Alignment Program (TMAP)
The Torrent Mapping Alignment Program (TMAP) is an alignment software for both short and long read sequences. It applies similar strategies like BWA, but can deal particularly well with Ion Torrent sequencing data as the algorithms address issues related to that type of sequencing data.
Isaac Alignment
The Isaac aligner is targeted at, but not limited to, read sequences produced by the Illumina HiSeq platform. The aligner uses a novel indexing scheme for the reference genome and efficiently uses CPU capabilities.
Bowtie
Bowtie comprises two alignment algorithms, Bowtie 1 and 2, for read sequence data. Bowtie 1 aligns short reads and indexes the reference genome by applying Burrows-Wheeler transform like BWA. Bowtie 2 is generally faster, more sensitive, and uses less memory than Bowtie 1 for reads longer than 50 bp. In addition, Bowtie 2 supports gapped alignment, i.e. can identify indels.
In-Memory Alignment Server
In-Memory Alignment Server is an alignment tool that is directly integrated and running within our in-memory computing platform. It uses the available main memory for faster index structures, can directly access native database operations, and is highly parallelized to exploit all computational resources. We provide this alignment algorithm in both our Single-Read and Paired-Read pipelines.
SAMtools and BcfTools
SAMtools is a software package with various functionality to manipulate read alignments, e.g. transformation from SAM into compressed BAM format and vice versa, sorting, merging, and indexing. BCFtools processes variant calls in the Variant Call Format (VCF) and its binary counterpart BCF. SAMtools and BCFtools can be combined to conduct variant calling, which is applied in our single-read pipelines.
Genome Analysis Toolkit (GATK)
The Genome Analysis Toolkit (GATK) is a software package offering a range of tools for the efficient processing of sequencing data. Its main focus lies on variant discovery and improving data quality for that step, e.g. by applying local realignment or base quality score recalibration. The tools are highly parallelized to efficiently process input data.
Picard Tools
Picard is a set of Java-based tools to manipulate SAM and BAM files. In our pipelines, we use it particularly for read duplicate detection.
